library(tidyverse); library(gtsummary)
library(performance)
pen <- palmerpenguins::penguinsHW 08: Multiple Linear Regression Modeling
Where most statistical modeling begins
Purpose
Fit a multiple regression model, assess model fit and interpret predictors of different data types.
Instructions
You will be fitting two linear regression models in this assignment. Both must have a quantitative predictor. The first will additionally have a binary predictor and the second model will have a categorical predictor containing more than 2 levels.
For each model you will do the following steps. Examples of each are provided below.
- Identify variables in a sentence and variable names.
- Write the mathematical model, defining each of your variables. For the binary and categorical variables you must include what each level of the variable means, indicating which one is the reference group. Double check this after you fit the model. If your reference group in the model is not the one stated here you will loose points.
- Fit the multivariable model
- Interpret all regression coefficients in a sentence that contains the point estimate
- Graphically assess the model fit and comment on whether or not the assumptions of a regression model are upheld.
Submission instructions
- Use the template provided: [QMD]
- Right click and ‘save as’, then upload this file into your
scriptsfolder in Posit Cloud.
- Right click and ‘save as’, then upload this file into your
- Upload your PDF to Canvas by the due date
Part 1: Multiple Linear Regression with one quantitative and one binary predictor.
1. Identify variables
- Quantitative outcome: Body Mass
- Quantitative predictor: Flipper length
- Binary predictor: Sex. This is an indicator of the penguin being male.
2. The mathematical model would look like:
- Let
body_mass_g - Let
flipper_length_mm - Let
sex='female' - Let
sex='male'
3. Fit the multivariable model
pen.sex.model <- lm(body_mass_g ~ flipper_length_mm + sex, data=pen)
tbl_regression(pen.sex.model, intercept = TRUE) %>%
add_glance_table(include = c("adj.r.squared", "nobs"))| Characteristic | Beta | 95% CI1 | p-value |
|---|---|---|---|
| (Intercept) | -5,410 | -5,973, -4,848 | <0.001 |
| flipper_length_mm | 47 | 44, 50 | <0.001 |
| sex | |||
| female | — | — | |
| male | 348 | 268, 427 | <0.001 |
| Adjusted R² | 0.805 | ||
| No. Obs. | 333 | ||
| 1 CI = Confidence Interval | |||
4. Interpret all regression coefficients of the multivariable model.
sex=0) who has no flippers (flipper_length_mm=0), their predicted average body mass is -5410g (95% CI -5973, -4848)
5. Assess model fit
check_normality(pen.sex.model) |> plot(type = "density")
check_normality(pen.sex.model) |> plot(type = "qq") +
scale_y_continuous(limits = c(-3, 3))
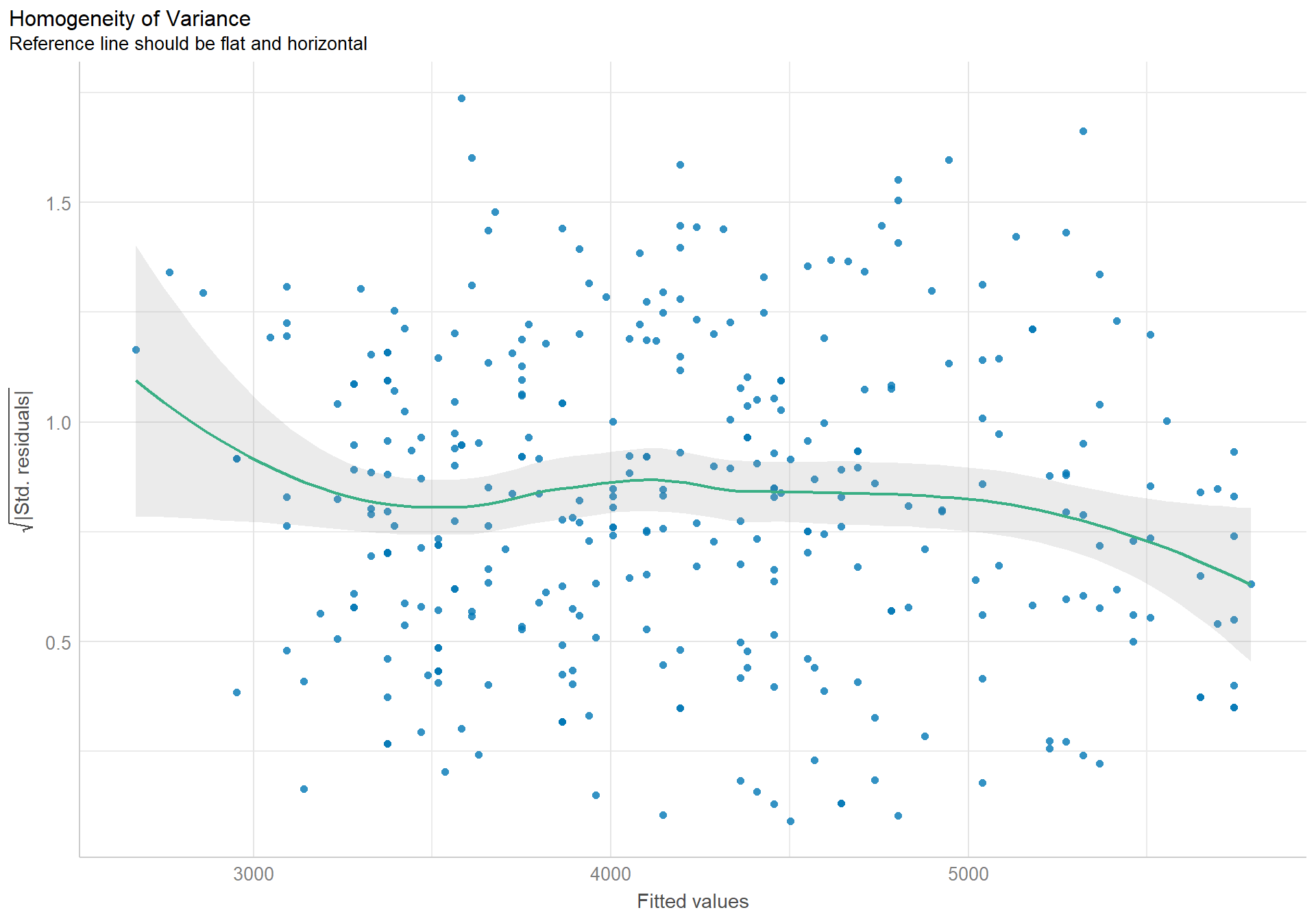
check_heteroscedasticity(pen.sex.model) |> plot() 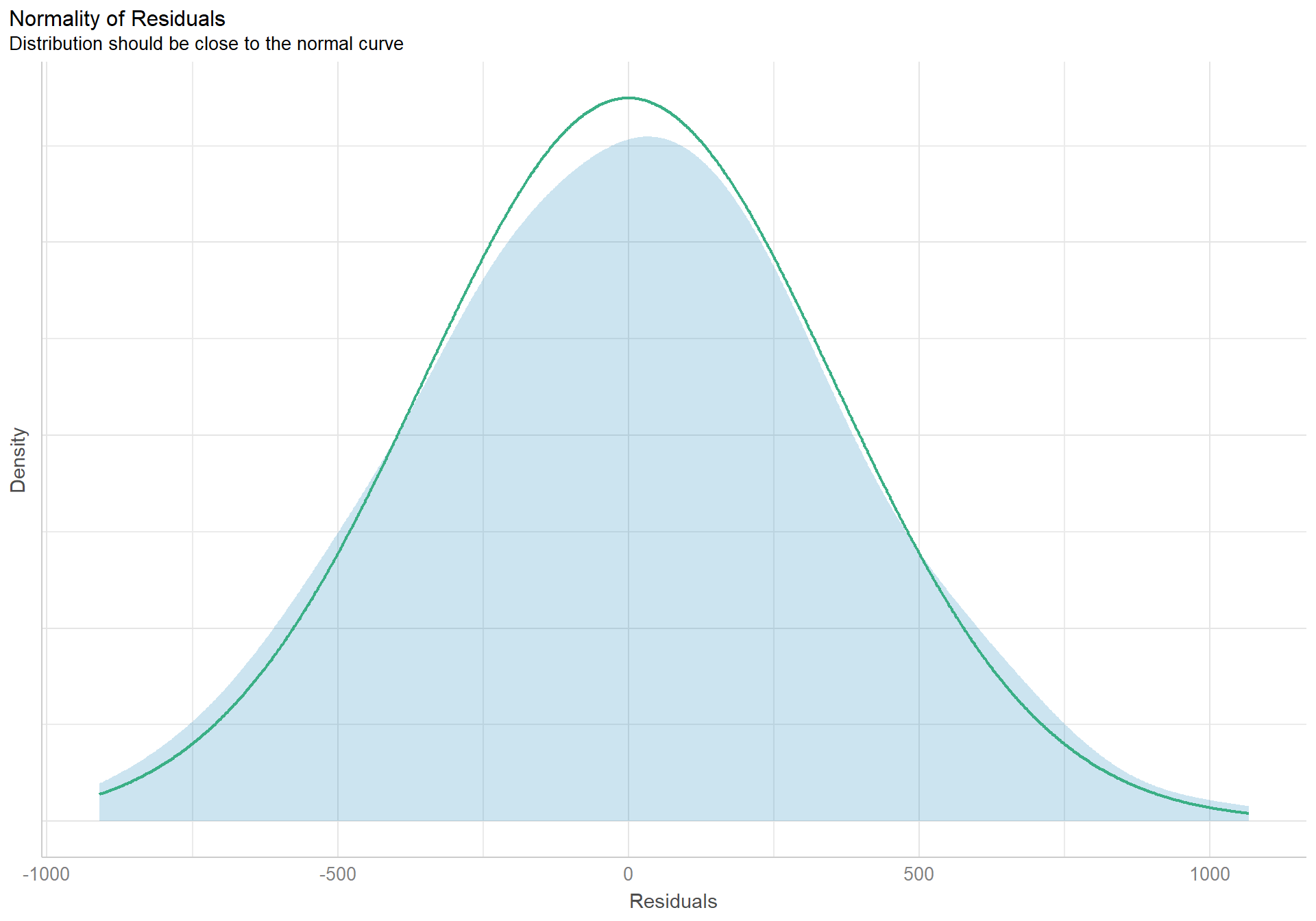
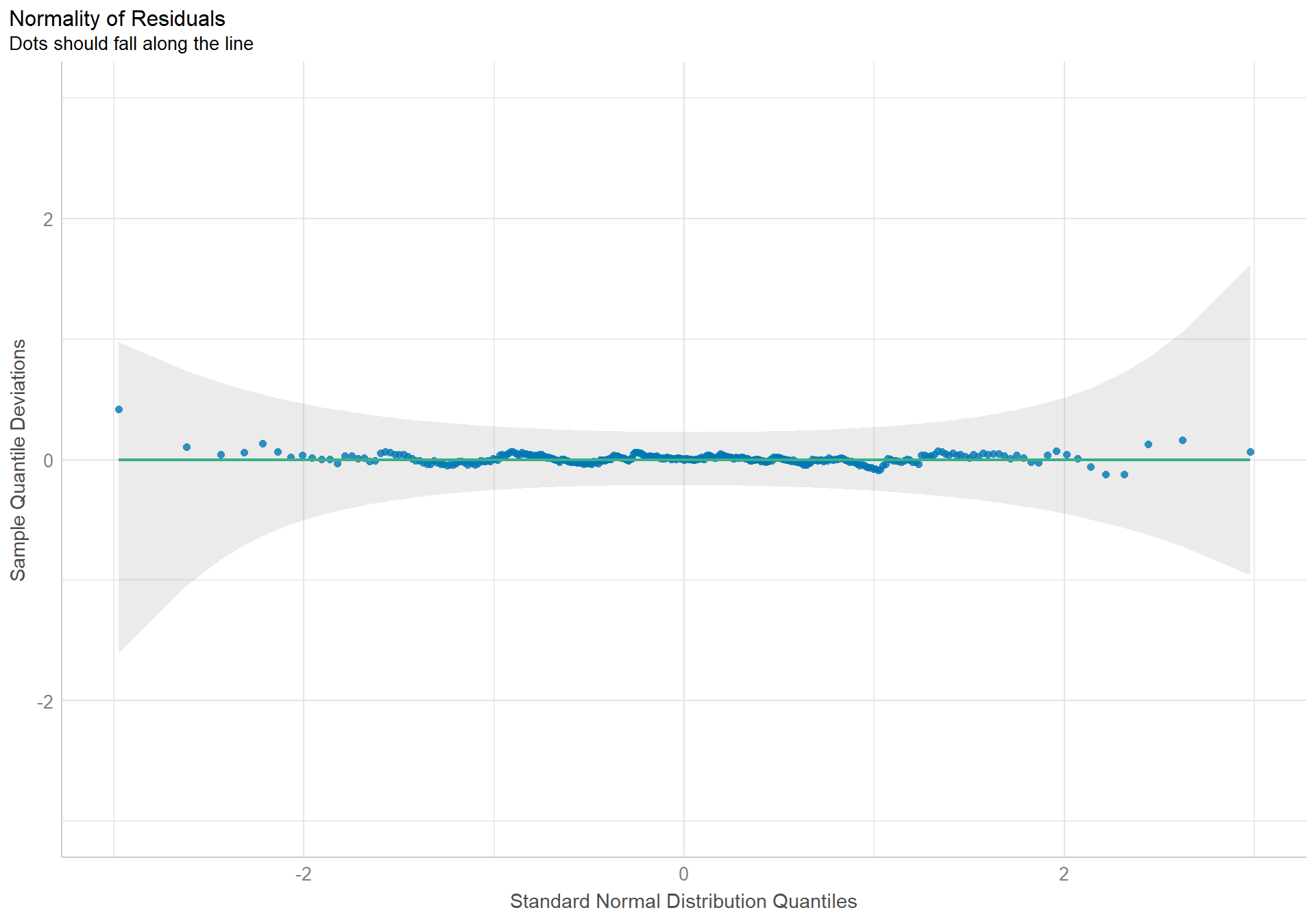
The density plot of the residuals looks like a nice pretty normal distribution, and the dots in the qqplot follow the reference line well.
The assumption of constant variance is also well enough upheld. The spike upward at the lower end of fitted values is being pulled up by a few outliers only, and the dip downward at the upper tail are all within normal amounts of variation.
Part 2: Categorical Predictor
1. Identify variables
- Quantitative outcome: Body Mass
- Quantitative predictor: Flipper length
- Binary predictor: Island.
2. The mathematical model would look like:
- Let
body_mass_g - Let
flipper_length_mm - Let
island='Dream'and 0 otherwise - Let
island='Torgersen'and 0 otherwise
The reference group for Island is Biscoe.
3. Fit the multivariable model
pen.island.model <- lm(body_mass_g ~ flipper_length_mm + island, data=pen)
tbl_regression(pen.island.model, intercept = TRUE) %>%
add_glance_table(include = c("adj.r.squared", "nobs"))| Characteristic | Beta | 95% CI1 | p-value |
|---|---|---|---|
| (Intercept) | -4,625 | -5,397, -3,853 | <0.001 |
| flipper_length_mm | 45 | 41, 48 | <0.001 |
| island | |||
| Biscoe | — | — | |
| Dream | -262 | -370, -154 | <0.001 |
| Torgersen | -185 | -323, -47 | 0.009 |
| Adjusted R² | 0.772 | ||
| No. Obs. | 342 | ||
| 1 CI = Confidence Interval | |||
4. Interpret all regression coefficients of the multivariable model.
5. Assess model fit
check_normality(pen.island.model) |> plot(type = "density")
check_normality(pen.island.model) |> plot(type = "qq") +
scale_y_continuous(limits = c(-3, 3))
check_heteroscedasticity(pen.island.model) |> plot() 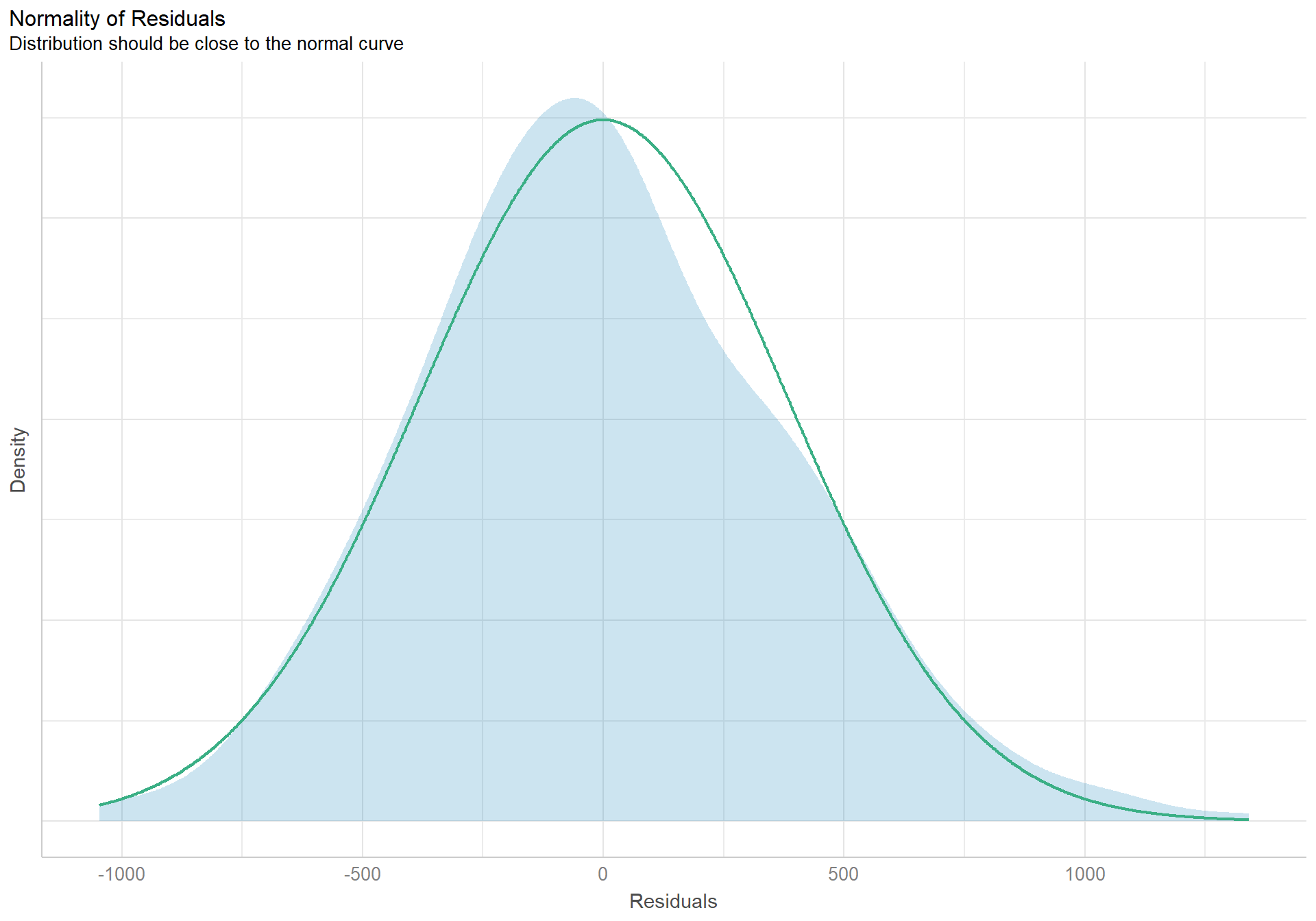
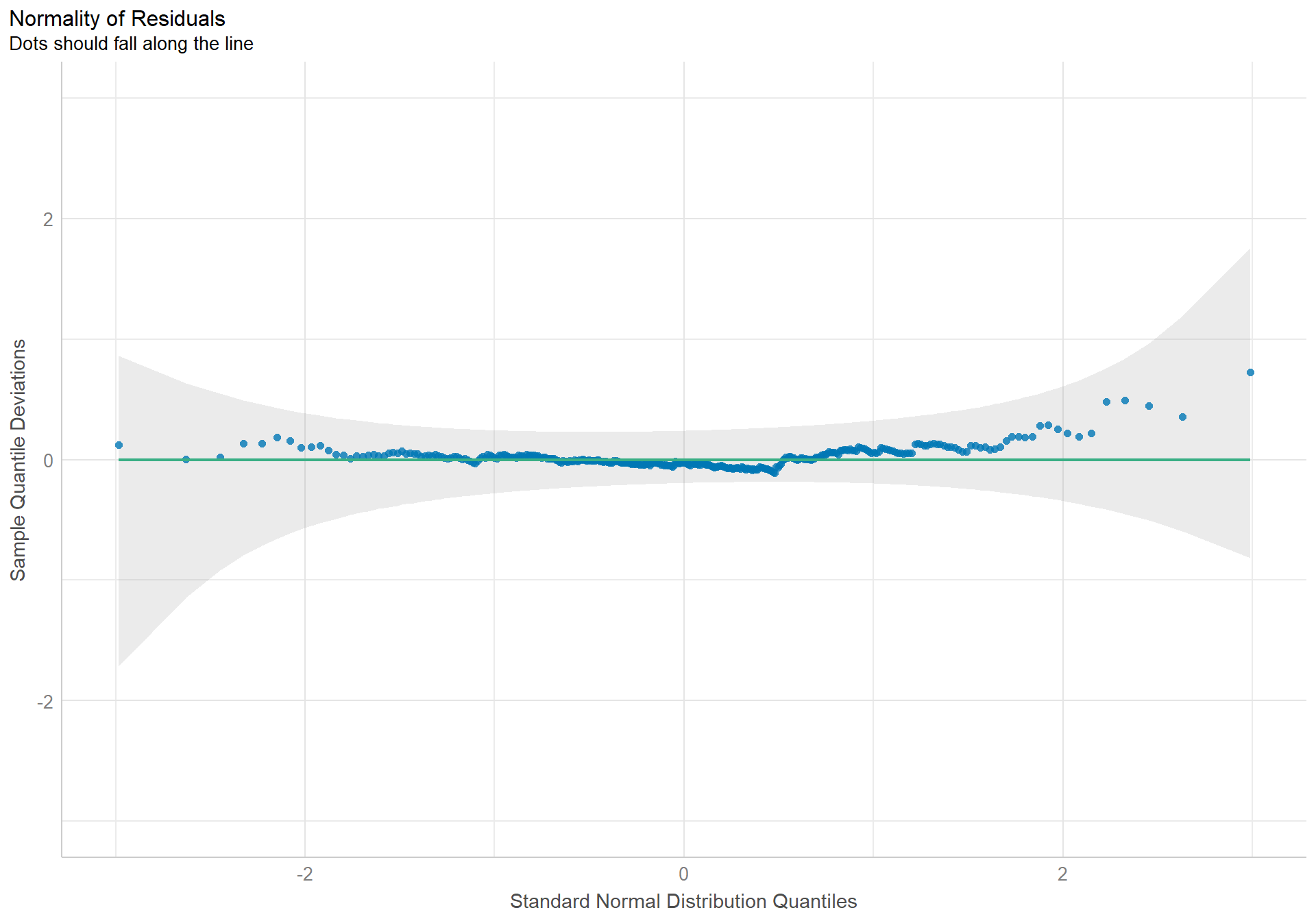
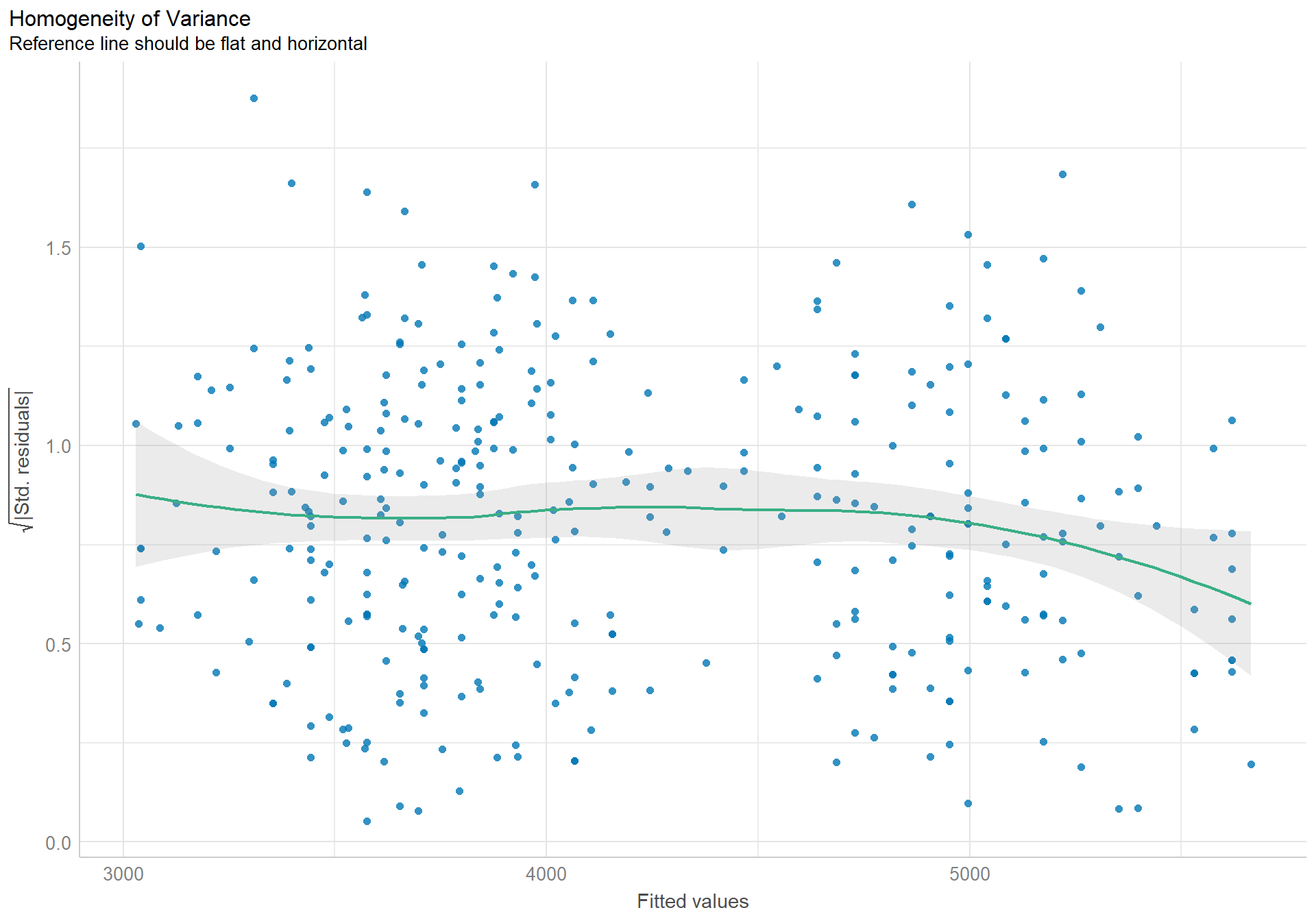
The density plot of the residuals looks like a nice pretty normal distribution, and the dots in the qqplot follow the reference line well.
The assumption of constant variance is also well enough upheld. The slight curved patter is within normal amounts of variation.